Unlocking the Secrets of Quantitative Disease Resistance
Temporal Regulation of WRKY6 in Solanum pennellii
Quantitative Disease Resistance (QDR) provides durable, broad-spectrum protection against pathogens like Sclerotinia sclerotiorum, a necrotrophic fungus infecting over 400 plant species. Unlike qualitative resistance, QDR is polygenic, offering partial but sustainable resistance under field conditions. Wild tomato relatives, such as Solanum pennellii, exhibit natural variation in QDR, making them ideal models for studying the regulatory timing of defense responses. While genetic loci associated with QDR have been identified, the temporal dynamics of gene expression during early infection remain poorly understood. Einspanier et al. (2025) investigated how early activation of defense genes, particularly the transcription factor WRKY6, influences QDR in S. pennellii challenged by S. sclerotiorum.
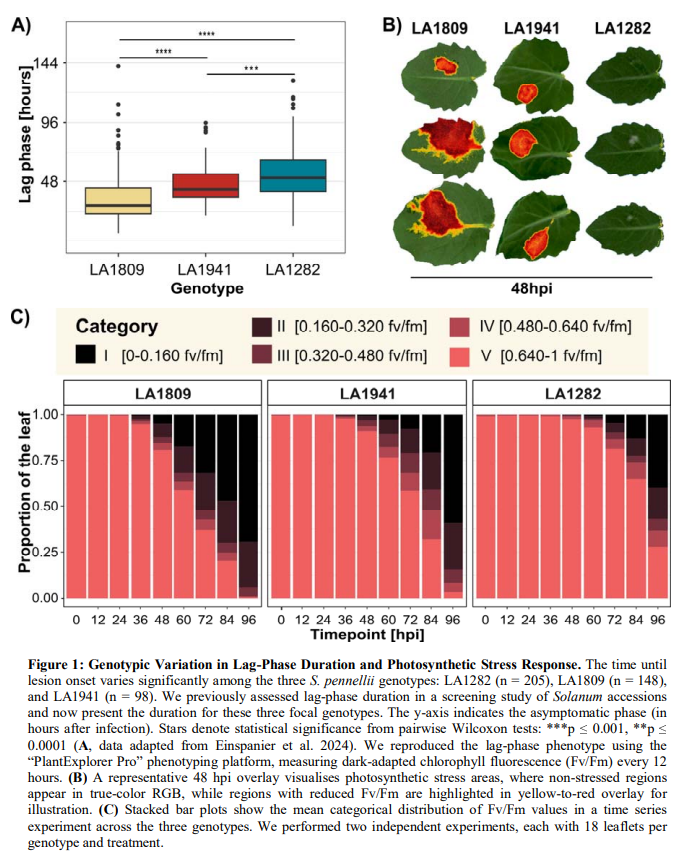
Three genetically distinct accessions of the wild tomato relative Solanum pennellii were selected based on their contrasting lag phase durations, making them ideal models for dissecting the temporal dynamics of defense responses:
- LA1282: Exhibits a prolonged lag phase (56.88 hours), representing a high-QDR genotype.
- LA1941: Displays an intermediate lag phase (49.92 hours), serving as a moderate-QDR genotype.
- LA1809: Characterized by a short lag phase (38.16 hours), representing a low-QDR genotype.

Test inoculum was prepared as a standardized fungal suspension of Sclerotinia sclerotiorum isolate 1980, cultivated from sclerotia on Potato Dextrose Agar (PDA) and incubated in Potato Dextrose Broth (PDB). The suspension was adjusted to an optical density (OD₆₀₀) of 1.0 and supplemented with Tween-80 to ensure uniform dispersion. Small droplets (10 μL) of the fungal suspension were applied to the center of detached Solanum pennellii leaves. Mock treatments consisted of PDB without fungal material, serving as negative controls.
Plants were cultivated under controlled environmental conditions (21°C, 65% relative humidity, 16-hour photoperiod) to ensure reproducibility and minimize environmental variability. This setup allowed for precise monitoring of disease progression and gene expression dynamics in response to S. sclerotiorum infection. The screening system consisted of a PlantExplorer Pro+, providing high-resolution Fv/Fm (chlorophyll fluorescence) imaging to monitor photosynthetic stress every 12 hours post-inoculation (hpi), and loss of photosynthetic efficiency as an indicator of pathogen impact. A Navautron Automated Lesion Tracker was used to record lesion expansion and to track disease progression.
Resistant genotypes (e.g., LA1282) exhibited a prolonged lag phase (56.88 hours), delayed photosynthetic stress, and sustained Fv/Fm values, while susceptible genotypes (e.g., LA1809) showed rapid lesion expansion and early decline in photosynthetic efficiency. Transcriptomic analysis identified 71 differentially expressed genes (DEGs) in the resistant genotype at 48 hours post-inoculation (hpi),
including pathogenesis-related (PR) genes, oxidative stress response genes (e.g., DOX1, LOX1), and transcription factors (e.g., WRKY6). WRKY6 emerged as a central regulator, with elevated basal expression in resistant genotypes, suggesting a primed defense state. Gene Set Enrichment Analysis (GSEA) further confirmed the activation of defense-related processes.
Conclusion
The study demonstrates that quantitative disease resistance (QDR) in Solanum pennellii against Sclerotinia sclerotiorum is driven by temporal shifts in gene expression, particularly during the lag phase of infection. The researchers identified WRKY6 as a central regulator of defense-related genes, with elevated basal expression in resistant genotypes contributing to delayed lesion onset. These findings underscore the importance of early defense activation and genotype-specific regulatory dynamics in shaping QDR, ultimately enhancing the plant’s ability to delay pathogen invasion.
For further details, please read the original publication:
- Einspanier, S., et al. (2025). Temporal Shifts in Gene Expression Drive Quantitative Resistance to a Necrotrophic Fungus in a Tomato Crop Wild Relative. BioRxiv. DOI: 10.1101/2025.01.01.522345



